Code
from pathlib import Path
from PIL import ImageJanuary 9, 2021
I want to try entering more kaggle competitions. There is a nice one available which is to segment images to spot specific functional groups of cells in images of the kidney. The images are very large so some means of processing smaller parts would be good.
I’ve started off the download for the images.
The challenge is a segmentation challenge. I need to create something that can spot the areas of the image that have the specific cell groups within them.
What I need is a way to turn the images into smaller images that I can then process, and then a way to turn the smaller images back into the full segmentation. This will allow me to use a pre-existing model to process the images and produce the segmentation.
It would be nice to have a way to scan the image at a high level to find areas of interest which can then be further processed.
The images are huge, which is not surprising. I want to look at them however I need to be able to cut a small section of the image out before viewing it.
I considered using image magik to cut the image up into parts. While this is possible it is quite slow, even with the smallest image.
I do have enough memory on this machine to load the images so I will just have to use the PIL library to cut them up. Lets try to do that now.
[PosixPath('/data/kaggle/HuBMAP/train/cb2d976f4.tiff'),
PosixPath('/data/kaggle/HuBMAP/train/0486052bb.tiff'),
PosixPath('/data/kaggle/HuBMAP/train/54f2eec69.tiff'),
PosixPath('/data/kaggle/HuBMAP/train/e79de561c.tiff'),
PosixPath('/data/kaggle/HuBMAP/train/2f6ecfcdf.tiff'),
PosixPath('/data/kaggle/HuBMAP/train/095bf7a1f.tiff'),
PosixPath('/data/kaggle/HuBMAP/train/aaa6a05cc.tiff'),
PosixPath('/data/kaggle/HuBMAP/train/1e2425f28.tiff')]DecompressionBombError: Image size (1731207120 pixels) exceeds limit of 178956970 pixels, could be decompression bomb DOS attack.That loaded the image … extremely fast. I’m guessing that just read the metadata and that the full image has not been loaded. Indeed this jupyter lab session isn’t even the largest one running on this machine.
That image looks really nice. It feels good to work on something that could really help medicine or the understanding of the human body.
What I want now is to be able to load the segmentations for this 1000x1000 image, or for any arbitrary region of an image.
Loading this image has used around 6 GB of memory or so. I must now assume that it has loaded the full image into memory. The size on disk is around 2.5 GB so I’m going to work with an assumption that the images more than double in size when loaded.
I have around 48 GB free right now (even with the above image) so I can load the training set into memory.
148 µs ± 988 ns per loop (mean ± std. dev. of 7 runs, 10000 loops each)So it’s only slow when loading the image from disk. The cropping operation is very fast.
That’s great as it means I can be quite aggressive with the cropping.
Now I want to investigate the features that are provided for this. The two json files seem to have a common structure.
(True, True){'type': 'Feature',
'id': 'PathAnnotationObject',
'geometry': {'type': 'Polygon',
'coordinates': [[[3562, 18742],
[3475, 18762],
[3451, 18826],
[3447, 18873],
[3502, 18941],
[3570, 18947],
[3612, 18923],
[3665, 18871],
[3672, 18801],
[3646, 18745],
[3562, 18742]]]},
'properties': {'classification': {'name': 'glomerulus', 'colorRGB': -3140401},
'isLocked': True,
'measurements': []}}Lets try seeing this glomerulus.
[[3562, 18742],
[3475, 18762],
[3451, 18826],
[3447, 18873],
[3502, 18941],
[3570, 18947],
[3612, 18923],
[3665, 18871],
[3672, 18801],
[3646, 18745],
[3562, 18742]]from typing import List
from PIL import ImageDraw
def crop_with_polygon(image: Image.Image, polygon: List[List[int]], margin: int = 10) -> Image.Image:
min_x, max_x = min(x for x, y in polygon), max(x for x, y in polygon)
min_y, max_y = min(y for x, y in polygon), max(y for x, y in polygon)
cropped_image = image.crop((min_x - margin, min_y - margin, max_x + margin, max_y + margin))
polygon_image = Image.new("RGBA", (max_x - min_x, max_y - min_y))
polygon_draw = ImageDraw.Draw(polygon_image)
polygon_draw.polygon([
(x-min_x, y-min_y)
for x, y in polygon
], fill=(255,255,255,127), outline=(255,255,255,255))
cropped_image.paste(polygon_image, (margin, margin), mask=polygon_image)
return cropped_imageThat’s actually much bigger than I was thinking before doing this cropping and projecting. The documentation for this task describes this as being the collection of cells that are centered around a capillary where every cell is within diffusion range of every other cell.
So I guess there is a capiliary near the center of that polygon. The diffusion requirement also suggests a reasonably fixed size. Lets try projecting a few of them.
from typing import Dict, Any, Tuple
Offset = Tuple[int, int]
Size = Tuple[int, int]
ImageWithOffsetAndSize = Tuple[Image.Image, Offset, Size]
Color = Tuple[int, int, int, int]
COLOR_SOLID_WHITE = (255,255,255,255)
COLOR_TRANSLUCENT_WHITE = (255,255,255,127)
def crop_and_draw(image: Image.Image, features: List[Dict[str, Any]], margin: int = 10) -> Image.Image:
cropped_image, offset, _ = crop_to_features(image, features)
draw_features(cropped_image, features, offset)
return cropped_image
def crop_to_features(image: Image.Image, features: List[Dict[str, Any]], margin: int = 10) -> ImageWithOffsetAndSize:
(polygon_offset_x, polygon_offset_y), (polygon_width, polygon_height) = polygon_offset_and_size([
point
for feature in features
for polygon in feature["geometry"]["coordinates"]
for point in polygon
])
assert polygon_width < 1024, f"width of {polygon_width:,} is too large"
assert polygon_height < 1024, f"height of {polygon_height:,} is too large"
cropped_image = image.crop((
polygon_offset_x - margin,
polygon_offset_y - margin,
polygon_offset_x + polygon_width + margin,
polygon_offset_y + polygon_height + margin,
))
return (
cropped_image,
(polygon_offset_x - margin, polygon_offset_y - margin),
(polygon_width + (margin * 2), polygon_height + (margin * 2)),
)
def polygon_offset_and_size(polygon: List[List[int]]) -> Tuple[Offset, Size]:
min_x, max_x = min(x for x, y in polygon), max(x for x, y in polygon)
min_y, max_y = min(y for x, y in polygon), max(y for x, y in polygon)
return ((min_x, min_y), (max_x - min_x, max_y - min_y))
def draw_features(
image: Image.Image,
features: List[Dict[str, Any]],
offset: Offset,
border: Color = COLOR_SOLID_WHITE,
fill: Color = COLOR_TRANSLUCENT_WHITE
) -> None:
for feature in features:
for polygon in feature["geometry"]["coordinates"]:
draw_polygon(image, polygon, offset, border=border, fill=fill)
def draw_polygon(
image: Image.Image,
polygon: List[List[int]],
offset: Offset,
border: Color = COLOR_SOLID_WHITE,
fill: Color = COLOR_TRANSLUCENT_WHITE
) -> None:
(polygon_offset_x, polygon_offset_y), polygon_size = polygon_offset_and_size(polygon)
polygon_image = Image.new("RGBA", polygon_size)
polygon_draw = ImageDraw.Draw(polygon_image)
polygon_draw.polygon([
(x-polygon_offset_x, y-polygon_offset_y)
for x, y in polygon
], fill=fill, outline=border)
image.paste(polygon_image, (polygon_offset_x - offset[0], polygon_offset_y - offset[1]), mask=polygon_image)The solution for this competition needs to produce a segmentation image which is just a mask of all anatomical structures. So I should check that this works with the anatomical version of the file as well.
I’m glad that I don’t have to recreate a polygon from a regular pixel level segmentation.
AssertionError: width of 20,038 is too largeThe other structures are way too big to reasonably draw on these, so to show them nicely I should either scale the image or just draw part of the structures. I’m going to try scaling the entire image after drawing every structure.
COLOR_SOLID_GREEN = (127,255,127,255)
COLOR_TRANSLUCENT_GREEN = (127,255,127,127)
def draw_everything(image: Image.Image, glomeruli: List[Dict[str, Any]], other_structures: List[Dict[str, Any]]) -> Image.Image:
image = image.copy()
for feature in other_structures:
for polygon in feature["geometry"]["coordinates"]:
draw_polygon(image, polygon, offset=(0,0), border=COLOR_SOLID_GREEN, fill=COLOR_TRANSLUCENT_GREEN)
for feature in glomeruli:
for polygon in feature["geometry"]["coordinates"]:
draw_polygon(image, polygon, offset=(0,0), border=COLOR_SOLID_WHITE, fill=COLOR_TRANSLUCENT_WHITE)
maxsize = (1028, 1028)
image.thumbnail(maxsize, Image.ANTIALIAS)
return image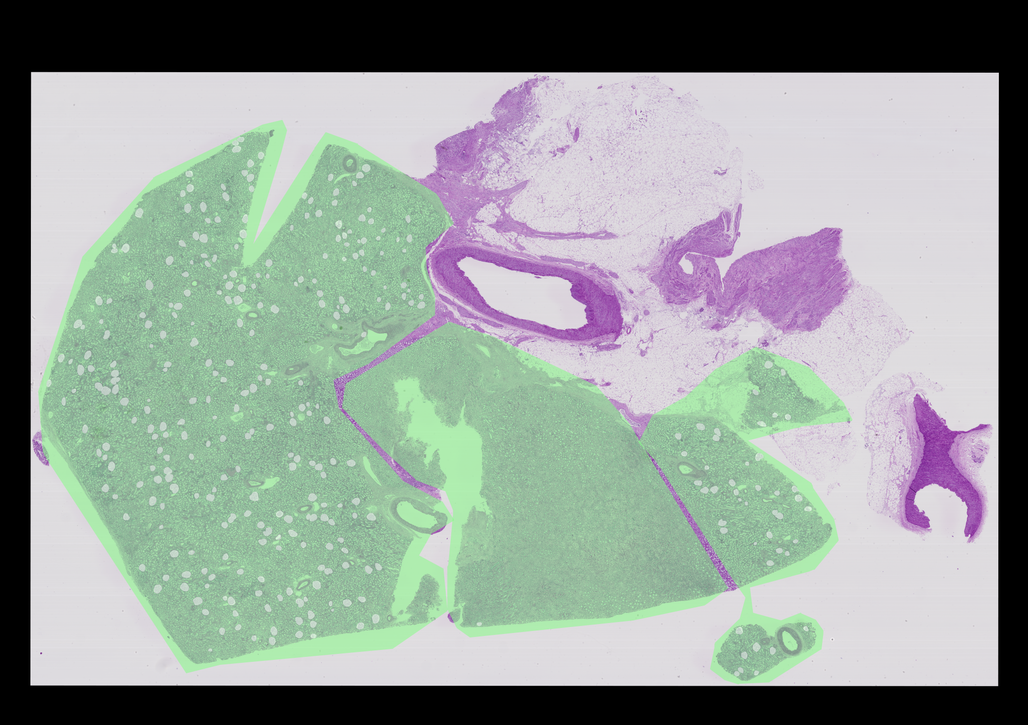
So the other features are so massive compared to the glomeruli, and that the glomeruli are only within these larger structures. This seems like quite an odd thing.
Right now I think that I need to have several different detectors, one for the glomeruli and one for everything else at least. Lets see how prevalent the glomeruli are.
# this now scales
def crop_to_features(image: Image.Image, features: List[Dict[str, Any]], margin: int = 10) -> ImageWithOffsetAndSize:
(polygon_offset_x, polygon_offset_y), (polygon_width, polygon_height) = polygon_offset_and_size([
point
for feature in features
for polygon in feature["geometry"]["coordinates"]
for point in polygon
])
cropped_image = image.crop((
polygon_offset_x - margin,
polygon_offset_y - margin,
polygon_offset_x + polygon_width + margin,
polygon_offset_y + polygon_height + margin,
))
if polygon_width > 1024 or polygon_height > 1024:
cropped_image.thumbnail((1024, 1024), Image.ANTIALIAS)
return (
cropped_image,
(polygon_offset_x - margin, polygon_offset_y - margin),
(polygon_width + (margin * 2), polygon_height + (margin * 2)),
)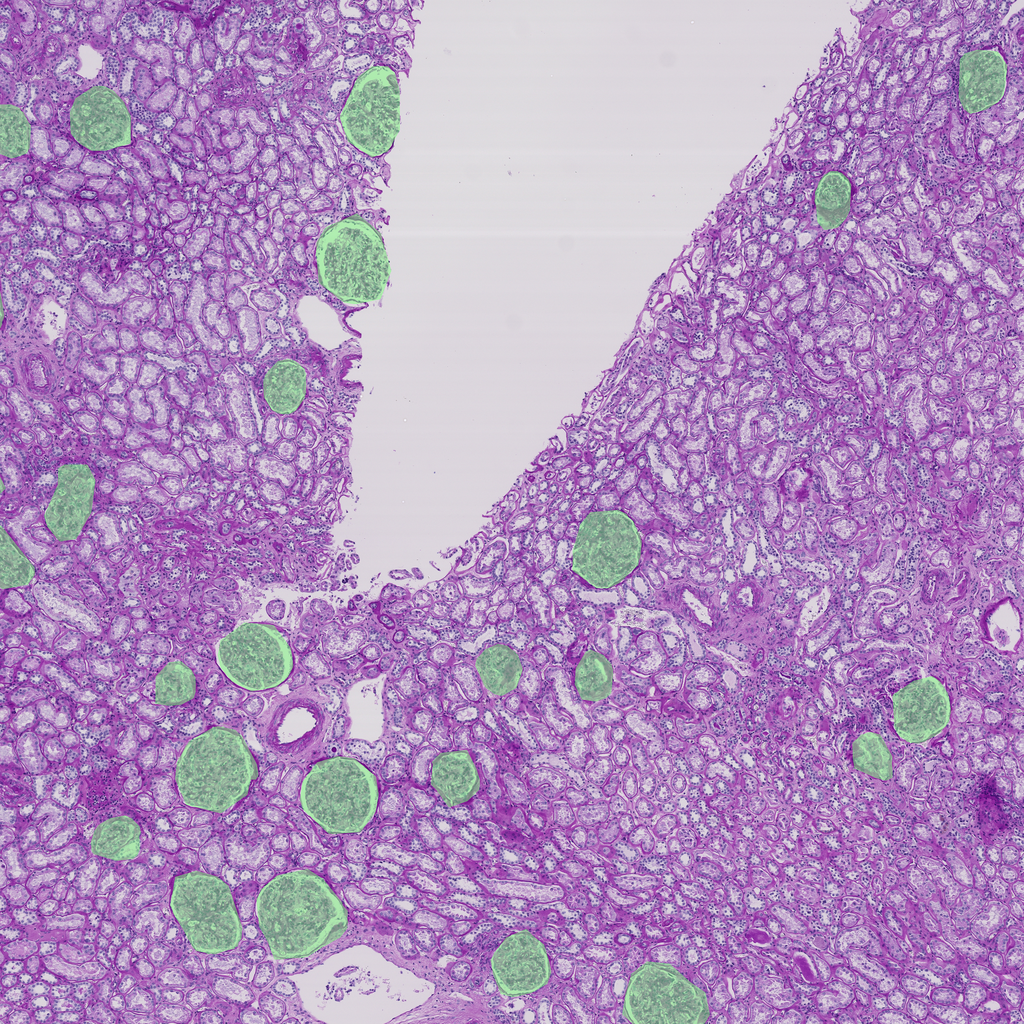
So the glomeruli are quite prevalent. This is a 5k x 5k area so it would be quite easy for a high resolution detector to hit an area that has no glomeruli.
My current thoughts are:
One concern I have is having enough training data to train the large scale anatomical structure segmenter. This will be trained from scratch and there are only 8 training images (so perhaps 24 structures). I might have to supplement that dataset - or find an existing segmenter for that, as the focus of this task is not the large scale structures.
I think I need to re-read those rules.
Your challenge is to detect functional tissue units (FTUs) across different tissue preparation pipelines. An FTU is defined as a “three-dimensional block of cells centered around a capillary, such that each cell in this block is within diffusion distance from any other cell in the same block” (de Bono, 2013). The goal of this competition is the implementation of a successful and robust glomeruli FTU detector.
So is the challenge just to find the glomeruli that lie within the larger anatomical structures? That means I don’t need the larger scale detector as the anatomical structure regions should be provided.
At this point this blog post has already become a monster. To keep things manageable I am going to perform the training in a separate post.