Code
from pathlib import Path
DATA_FOLDER = Path("/data/blog/2021-09-24-bird-brain/")
IMAGE_PDF = DATA_FOLDER / "images.pdf"
IMAGES_FOLDER = DATA_FOLDER / "images"
IMAGES_FOLDER.mkdir(parents=True, exist_ok=True)September 24, 2021
My friends know of my interest in deep learning and someone linked me a paper about using pigeons to classify mammograms to identify breast cancer (Levenson 2015). When I saw this I immediately wanted to recreate this test using a computer vision model. The trained pigeon accuracy reaches 85% after 15 days - can I do better than that?
The dataset was 144 images at three levels of magnification. This is quite a small number for training a model from an imagenet base. It would be good to find a model that has been pretrained on medical images.
The other problem with replicating this study is that the images are provided in pdf files. I have to extract them from that and ensure that they are correctly labelled.
Looking at the pdf there are 288 images that are split into set A and set B. Lets start by extracting the images and labelling them.
import fitz
pages = [
"4x-normal-a",
"4x-normal-b",
"4x-cancer-a",
"4x-cancer-b",
"10x-normal-a",
"10x-normal-b",
"10x-cancer-a",
"10x-cancer-b",
"20x-normal-a",
"20x-normal-b",
"20x-cancer-a",
"20x-cancer-b",
"10x-normal-monochrome-a",
"10x-normal-monochrome-b",
"10x-cancer-monochrome-a",
"10x-cancer-monochrome-b",
"10x-normal-monochrome-115-a",
"10x-normal-monochrome-115-b",
"10x-cancer-monochrome-115-a",
"10x-cancer-monochrome-115-b",
"10x-normal-monochrome-127-a",
"10x-normal-monochrome-127-b",
"10x-cancer-monochrome-127-a",
"10x-cancer-monochrome-127-b",
]
doc = fitz.open(IMAGE_PDF)
assert len(doc) == len(pages)
for page, name in enumerate(pages):
for index, image in enumerate(doc.getPageImageList(page)):
path = IMAGES_FOLDER / f"{name}-{index:02d}.png"
xref = image[0]
pix = fitz.Pixmap(doc, xref)
if pix.n >= 5: # CMYK: convert to RGB first
pix = fitz.Pixmap(fitz.csRGB, pix)
pix.writePNG(path)
pix = NoneWe can have a quick look at one to ensure it has loaded correctly:
Loaded 10x-cancer-a-00.png (288, 288)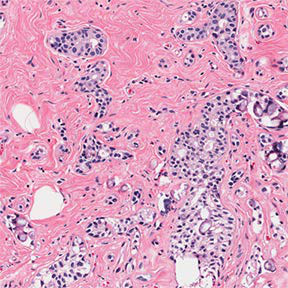
This can be seen as the first image on page 7, so it looks like the extraction has worked perfectly. Let’s check the image dimensions.
{(288, 287), (288, 288), (288, 294), (384, 384)}These are probably close enough? I think that the bigger problem is the variance in zoom level.
I want to create a databunch out of these images. I can tell from the path what the label is so it might well be possible to just create the databunch with one of the builder methods.
The next thing is to create a labelled df from this.
from fastai.vision.all import (
ImageDataLoaders,
aug_transforms,
Resize,
)
def image_label(path: str) -> int:
return "cancer" if "cancer" in path else "normal"
dls = ImageDataLoaders.from_name_func(
path=IMAGES_FOLDER,
fnames=sorted(IMAGES_FOLDER.glob("*.png")),
label_func=image_label,
item_tfms=[Resize(224)], # 224 for resnet
batch_tfms=aug_transforms(
mult=1.0,
do_flip=True,
flip_vert=True,
max_rotate=10.0,
min_zoom=1.0,
max_zoom=1.1,
max_lighting=0.2,
max_warp=0., # all images are fixed perspective
p_affine=0.75,
p_lighting=0.75,
xtra_tfms=[Resize(224)],
size=None,
mode='bilinear',
pad_mode='reflection',
align_corners=True,
batch=False,
min_scale=1.0,
),
)Let’s give it a go.
/home/matthew/.cache/pypoetry/virtualenvs/blog-HrtMnrOS-py3.9/lib/python3.9/site-packages/fastai/callback/schedule.py:269: UserWarning: color is redundantly defined by the 'color' keyword argument and the fmt string "ro" (-> color='r'). The keyword argument will take precedence.
ax.plot(val, idx, 'ro', label=nm, c=color)
SuggestedLRs(valley=0.0008317637839354575)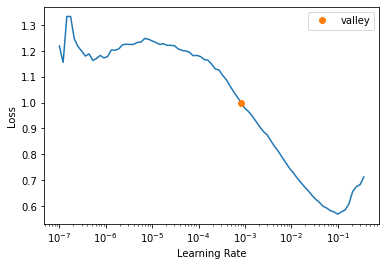
| epoch | train_loss | valid_loss | accuracy | error_rate | time |
|---|---|---|---|---|---|
| 0 | 1.084662 | 0.609927 | 0.771930 | 0.228070 | 00:02 |
| 1 | 1.015930 | 0.627861 | 0.736842 | 0.263158 | 00:02 |
| epoch | train_loss | valid_loss | accuracy | error_rate | time |
|---|---|---|---|---|---|
| 0 | 0.709828 | 0.471480 | 0.824561 | 0.175439 | 00:02 |
| 1 | 0.605629 | 0.371742 | 0.912281 | 0.087719 | 00:02 |
| 2 | 0.573627 | 0.277116 | 0.912281 | 0.087719 | 00:02 |
| 3 | 0.500686 | 0.233057 | 0.912281 | 0.087719 | 00:02 |
| 4 | 0.443576 | 0.211415 | 0.929825 | 0.070175 | 00:02 |
So this claims 92% accurcy. I could recreate the original data split to test if the data split was mean on the pigeons. Either way this seems pretty good?
I would never ship this though. The 8% error rate given the uneven distribution of classes (far more people are fine) means that most cancer predictions would be false positives.
So the original experiment has a bunch of refinement that means this original “replication” is rather poor. The changes in magnification level do not mean that there were associated changes in the source material - so image 1 at 4x resolution is the same as image 1 at 10x resolution.
You can clearly see that this is the same image at different levels of magnification. Since the dataset is split into A and B I can establish that there is no replication of the test set in the training set if I just use one side to train and the other to validate.
from fastai.vision.all import (
ImageDataLoaders,
aug_transforms,
Resize,
)
import torch
def image_label(path: str) -> int:
return "cancer" if "cancer" in path else "normal"
item_transforms = [Resize(224)] # 224 for resnet
batch_transforms = aug_transforms(
mult=1.0,
do_flip=True,
flip_vert=True,
max_rotate=10.0,
min_zoom=1.0,
max_zoom=1.1,
max_lighting=0.2,
max_warp=0., # all images are fixed perspective
p_affine=0.75,
p_lighting=0.75,
xtra_tfms=[Resize(224)],
size=None,
mode='bilinear',
pad_mode='reflection',
align_corners=True,
batch=False,
min_scale=1.0,
)
train_ds = ImageDataLoaders.from_name_func(
path=IMAGES_FOLDER,
fnames=sorted(IMAGES_FOLDER.glob("*-a-*.png")),
label_func=image_label,
item_tfms=item_transforms,
batch_tfms=batch_transforms,
valid_pct=0.,
).train
valid_ds = ImageDataLoaders.from_name_func(
path=IMAGES_FOLDER,
fnames=sorted(IMAGES_FOLDER.glob("*-b-*.png")),
label_func=image_label,
item_tfms=item_transforms,
batch_tfms=batch_transforms,
valid_pct=0.,
).train
dls = ImageDataLoaders(
train_ds,
valid_ds,
path=IMAGES_FOLDER,
device=torch.device("cuda")
)SuggestedLRs(valley=0.0012022644514217973)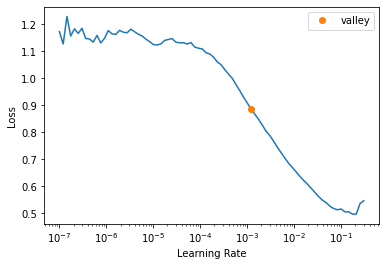
| epoch | train_loss | valid_loss | accuracy | error_rate | time |
|---|---|---|---|---|---|
| 0 | 1.210299 | 0.611843 | 0.648438 | 0.351562 | 00:02 |
| 1 | 1.056809 | 0.486527 | 0.796875 | 0.203125 | 00:02 |
| 2 | 0.943421 | 0.422140 | 0.835938 | 0.164062 | 00:02 |
| 3 | 0.842579 | 0.305688 | 0.875000 | 0.125000 | 00:02 |
| 4 | 0.759867 | 0.342617 | 0.867188 | 0.132812 | 00:02 |
| epoch | train_loss | valid_loss | accuracy | error_rate | time |
|---|---|---|---|---|---|
| 0 | 0.318117 | 0.338621 | 0.882812 | 0.117188 | 00:02 |
| 1 | 0.263593 | 0.223039 | 0.914062 | 0.085938 | 00:02 |
| 2 | 0.273602 | 0.352680 | 0.882812 | 0.117188 | 00:02 |
| 3 | 0.235429 | 0.256570 | 0.890625 | 0.109375 | 00:02 |
| 4 | 0.213778 | 0.328133 | 0.890625 | 0.109375 | 00:02 |
| 5 | 0.190749 | 0.325845 | 0.914062 | 0.085938 | 00:02 |
| 6 | 0.173745 | 0.304393 | 0.921875 | 0.078125 | 00:02 |
| 7 | 0.165102 | 0.269543 | 0.921875 | 0.078125 | 00:02 |
| 8 | 0.150334 | 0.298939 | 0.921875 | 0.078125 | 00:02 |
| 9 | 0.137877 | 0.309571 | 0.914062 | 0.085938 | 00:02 |
This gets to the same accuracy even with the good split approach. I had to train it a bit longer though. I think that the original training run could’ve gone for more epochs to get a higher peak performance.